news
Launch of an activity-based probe for the deubiquitinating enzyme OTULIN: UbiQ-121
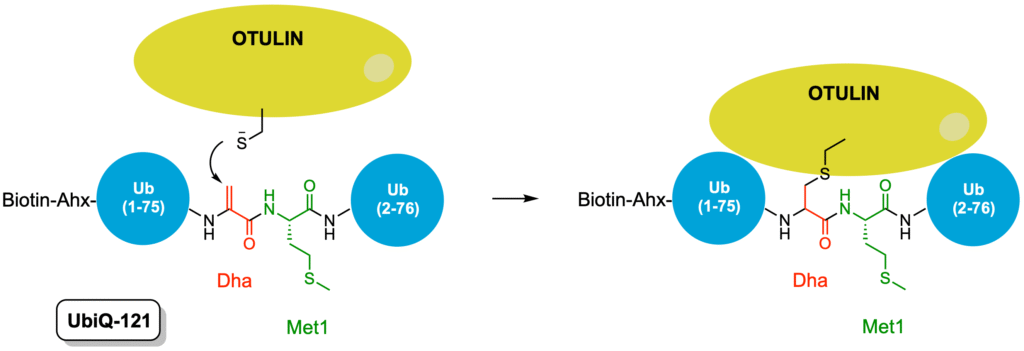
 Hereby we present UbiQ-121 (Figure 1) as an activity-based probe for OTULIN, a deubiquitinating enzyme that is specific for Met1-linked ubiquitin chains and acts as a negative regulator of nuclear factor κB signaling and immune homeostasis. In a collaborative effort published in Cell Chemical Biology, UbiQ-121 was designed and synthesised by UbiQ, evaluated for OTULIN biology by the group of Prof Daniel Krappmann (Helmholtz Zentrum Munich, Germany), evaluated by proteomics by the group of Prof Benedikt Kessler (Univ. of Oxford, UK) and structural biology experiments were performed by the group of Dr David Komander (MRC Lab of Mol. Biology, UK). As depicted in Figure 1, UbiQ-121 is based on Met1-linked diubiquitin in which G76 of the distal ubiquitin is replaced by an electrophilic dehydroalanine (Dha) residue. The catalytic cysteine residue of OTULIN reacts in a covalent manner with the Dha residue and as such UbiQ-121 captures OTULIN in its active conformation.
Hereby we present UbiQ-121 (Figure 1) as an activity-based probe for OTULIN, a deubiquitinating enzyme that is specific for Met1-linked ubiquitin chains and acts as a negative regulator of nuclear factor κB signaling and immune homeostasis. In a collaborative effort published in Cell Chemical Biology, UbiQ-121 was designed and synthesised by UbiQ, evaluated for OTULIN biology by the group of Prof Daniel Krappmann (Helmholtz Zentrum Munich, Germany), evaluated by proteomics by the group of Prof Benedikt Kessler (Univ. of Oxford, UK) and structural biology experiments were performed by the group of Dr David Komander (MRC Lab of Mol. Biology, UK). As depicted in Figure 1, UbiQ-121 is based on Met1-linked diubiquitin in which G76 of the distal ubiquitin is replaced by an electrophilic dehydroalanine (Dha) residue. The catalytic cysteine residue of OTULIN reacts in a covalent manner with the Dha residue and as such UbiQ-121 captures OTULIN in its active conformation.
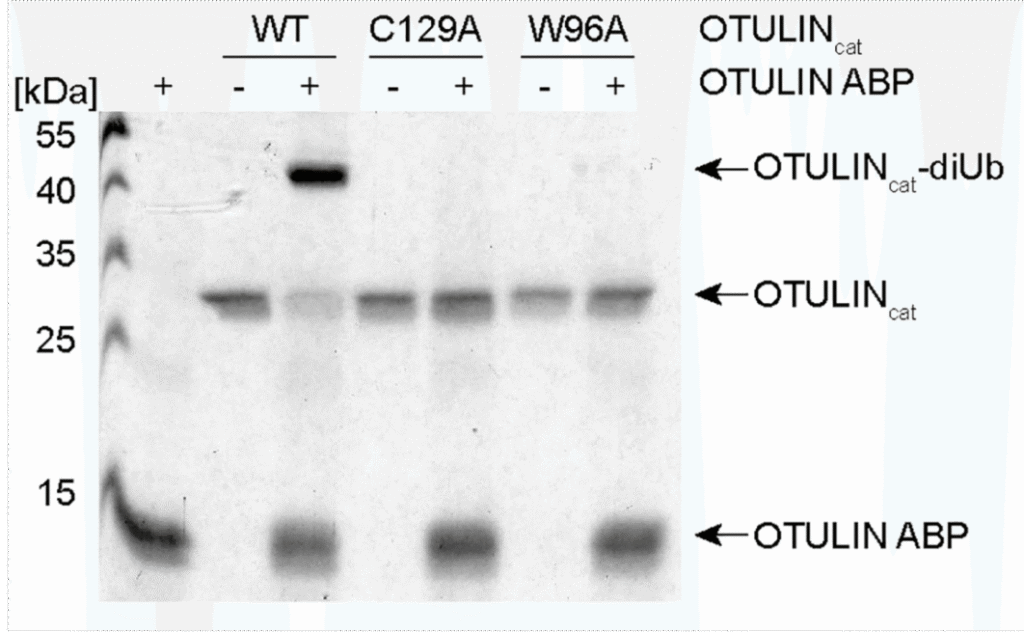
Figure 1. Left: Mode of action labeling of OTULIN by UbiQ-121. Right: recombinant OTULINcat (1 μg) was incubated with 4 μg UbiQ-121 (2 hr, 37°C). OTULIN-diUb complex formation was analyzed by Coomassie staining.